Hands-On with VDBBench: Benchmarking Vector Databases for POCs That Match Production
Vector databases are now a core part of AI infrastructure, powering various LLM-powered applications for customer service, content generation, search, recommendations, and more.
With so many options in the market, from purpose-built vector databases like Milvus and Zilliz Cloud to traditional databases with vector search as an add-on, choosing the right one isn’t as simple as reading benchmark charts.
Most teams run a Proof of Concept (POC) before committing, which is smart in theory — but in practice, many vendor benchmarks that look impressive on paper collapse under real-world conditions.
One of the main reasons is that most performance claims are based on outdated datasets from 2006–2012 (SIFT, GloVe, LAION) that behave very differently from modern embeddings. For example, SIFT uses 128-dimensional vectors, while today’s AI models produce far higher dimensions — 3,072 for OpenAI’s latest, 1,024 for Cohere’s — a big shift that impacts performance, cost, and scalability.
The Fix: Test with Your Data, Not Canned Benchmarks
The simplest and most effective solution: run your POC evaluation with the vectors your application actually generates. That means using your embedding models, your real queries, and your actual data distribution.
This is exactly what VDBBench — an open-source vector database benchmarking tool — is built for. It supports the evaluation and comparison of any vector database, including Milvus, Elasticsearch, pgvector, and more, and simulates real production workloads.
Download VDBBench 1.0 → | View Leaderboard → | What is VDBBench
VDBbench lets you:
Test with your own data from your embedding models
Simulate concurrent inserts, queries, and streaming ingestion
Measure P95/P99 latency, sustained throughput, and recall accuracy
Benchmark across multiple databases under identical conditions
Allows custom dataset testing so results actually match production
Next, we’ll walk you through how to run a production-grade POC with VDBBench and your real data — so you can make a confident, future-proof choice.
How to Evaluate VectorDBs with Your Custom Datasets with VDBBench
Before getting started, ensure you have Python 3.11 or higher installed. You’ll need vector data in CSV or NPY format, approximately 2-3 hours for complete setup and testing, and intermediate Python knowledge for troubleshooting if needed.
Installation and Configuration
If you’re evaluating one database, run this command:
pip install vectordb-bench
If you’re to compare all supported databases, run the command:
pip install vectordb-bench[all]
For specific database clients (eg: Elasticsearch):
pip install vectordb-bench[elastic]
Check this GitHub page for all the supported databases and their install commands.
Launching VDBBench
Start VDBBench with:
init_bench
Expected console output:
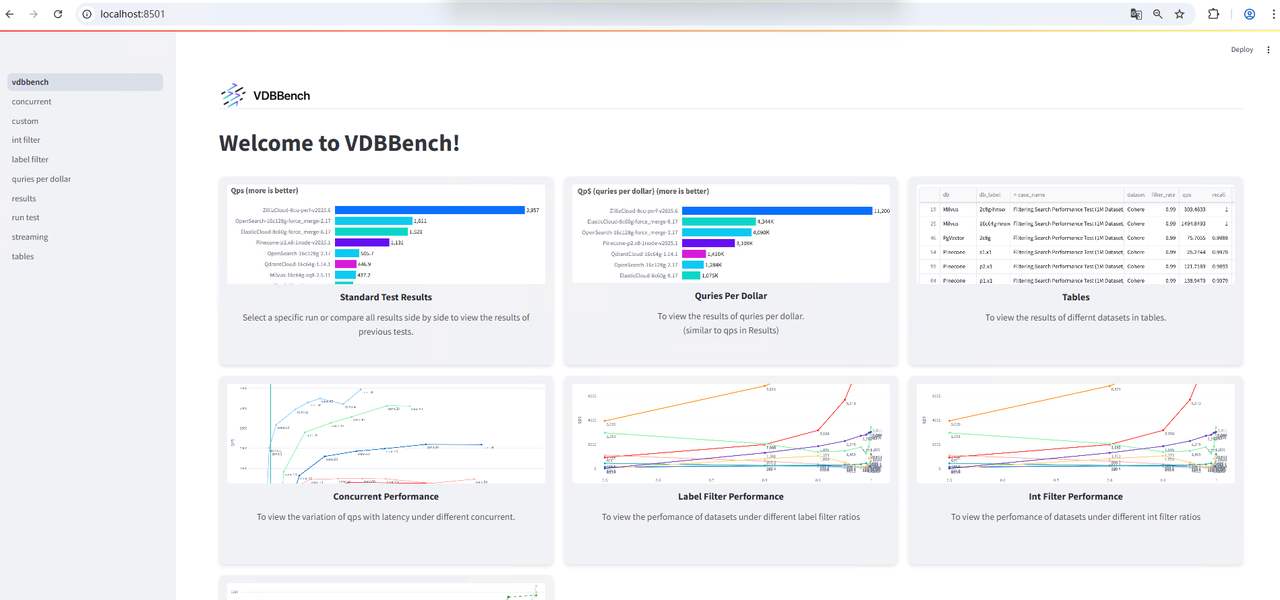
The web interface will be available locally:
Data Preparation and Format Conversion
VDBBench requires structured Parquet files with specific schemas to ensure consistent testing across different databases and datasets.
| File Name | Purpose | Required | Content Example |
|---|---|---|---|
| train.parquet | Vector collection for database insertion | ✅ | Vector ID + Vector data (list[float]) |
| test.parquet | Vector collection for queries | ✅ | Vector ID + Vector data (list[float]) |
| neighbors.parquet | Ground Truth for query vectors (actual nearest neighbor ID list) | ✅ | query_id -> [top_k similar ID list] |
| scalar_labels.parquet | Labels (metadata describing entities other than vectors) | ❌ | id -> label |
Required File Specifications:
Training Vector File (train.parquet) must contain an ID column with incremental integers and a vector column containing float32 arrays. Column names are configurable, but the ID column must use integer types for proper indexing.
Test Vector File (test.parquet) follows the same structure as the training data. The ID column name must be “id” while vector column names can be customized to match your data schema.
Ground Truth File (neighbors.parquet) contains the reference nearest neighbors for each test query. It requires an ID column corresponding to test vector IDs and a neighbors array column containing the correct nearest neighbor IDs from the training set.
Scalar Labels File (scalar_labels.parquet) is optional and contains metadata labels associated with training vectors, useful for filtered search testing.
Data Format Challenges
Most production vector data exists in formats that don’t directly match VDBBench requirements. CSV files typically store embeddings as string representations of arrays, NPY files contain raw numerical matrices without metadata, and database exports often use JSON or other structured formats.
Converting these formats manually involves several complex steps: parsing string representations into numerical arrays, computing exact nearest neighbors using libraries like FAISS, properly splitting datasets while maintaining ID consistency, and ensuring all data types match Parquet specifications.
Automated Format Conversion
To streamline the conversion process, we’ve developed a Python script that handles format conversion, ground truth computation, and proper data structuring automatically.
CSV Input Format:
id,emb,label
1,"[0.12,0.56,0.89,...]",A
2,"[0.33,0.48,0.90,...]",B
NPY Input Format:
import numpy as np
vectors = np.random.rand(10000, 768).astype('float32')
np.save("vectors.npy", vectors)
Conversion Script Implementation
Install required dependencies:
pip install numpy pandas faiss-cpu
Execute the conversion:
python convert_to_vdb_format.py \
--train data/train.csv \
--test data/test.csv \
--out datasets/custom \
--topk 10
Parameter Reference:
| Parameter Name | Required | Type | Description | Default Value |
|---|---|---|---|---|
--train | Yes | String | Training data path, supports CSV or NPY format. CSV must contain emb column, if no id column will auto-generate | None |
--test | Yes | String | Query data path, supports CSV or NPY format. Format same as training data | None |
--out | Yes | String | Output directory path, saves converted parquet files and neighbor index files | None |
--labels | No | String | Label CSV path, must contain labels column (formatted as string array), used for saving labels | None |
--topk | No | Integer | Number of nearest neighbors to return when computing | 10 |
Output Directory Structure:
datasets/custom/
├── train.parquet # Training vectors
├── test.parquet # Query vectors
├── neighbors.parquet # Ground Truth
└── scalar_labels.parquet # Optional scalar labels
Complete Conversion Script
import os
import argparse
import numpy as np
import pandas as pd
import faiss
from ast import literal_eval
from typing import Optional
def load_csv(path: str):
df = pd.read_csv(path)
if 'emb' not in df.columns:
raise ValueError(f"CSV file missing 'emb' column: {path}")
df['emb'] = df['emb'].apply(literal_eval)
if 'id' not in df.columns:
df.insert(0, 'id', range(len(df)))
return df
def load_npy(path: str):
arr = np.load(path)
df = pd.DataFrame({
'id': range(arr.shape[0]),
'emb': arr.tolist()
})
return df
def load_vectors(path: str) -> pd.DataFrame:
if path.endswith('.csv'):
return load_csv(path)
elif path.endswith('.npy'):
return load_npy(path)
else:
raise ValueError(f"Unsupported file format: {path}")
def compute_ground_truth(train_vectors: np.ndarray, test_vectors: np.ndarray, top_k: int = 10):
dim = train_vectors.shape[1]
index = faiss.IndexFlatL2(dim)
index.add(train_vectors)
_, indices = index.search(test_vectors, top_k)
return indices
def save_ground_truth(df_path: str, indices: np.ndarray):
df = pd.DataFrame({
"id": np.arange(indices.shape[0]),
"neighbors_id": indices.tolist()
})
df.to_parquet(df_path, index=False)
print(f"✅ Ground truth saved successfully: {df_path}")
def main(train_path: str, test_path: str, output_dir: str,
label_path: Optional[str] = None, top_k: int = 10):
os.makedirs(output_dir, exist_ok=True)
# Load training and query data
print("📥 Loading training data...")
train_df = load_vectors(train_path)
print("📥 Loading query data...")
test_df = load_vectors(test_path)
# Extract vectors and convert to numpy
train_vectors = np.array(train_df['emb'].to_list(), dtype='float32')
test_vectors = np.array(test_df['emb'].to_list(), dtype='float32')
# Save parquet files retaining all fields
train_df.to_parquet(os.path.join(output_dir, 'train.parquet'), index=False)
print(f"✅ train.parquet saved successfully, {len(train_df)} records total")
test_df.to_parquet(os.path.join(output_dir, 'test.parquet'), index=False)
print(f"✅ test.parquet saved successfully, {len(test_df)} records total")
# Compute ground truth
print("🔍 Computing Ground Truth (nearest neighbors)...")
gt_indices = compute_ground_truth(train_vectors, test_vectors, top_k=top_k)
save_ground_truth(os.path.join(output_dir, 'neighbors.parquet'), gt_indices)
# Load and save label file (if provided)
if label_path:
print("📥 Loading label file...")
label_df = pd.read_csv(label_path)
if 'labels' not in label_df.columns:
raise ValueError("Label file must contain 'labels' column")
label_df['labels'] = label_df['labels'].apply(literal_eval)
label_df.to_parquet(os.path.join(output_dir, 'scalar_labels.parquet'), index=False)
print("✅ Label file saved as scalar_labels.parquet")
if
name
== "__main__":
parser = argparse.ArgumentParser(description="Convert CSV/NPY vectors to VectorDBBench data format (retaining all columns)")
parser.add_argument("--train", required=True, help="Training data path (CSV or NPY)")
parser.add_argument("--test", required=True, help="Query data path (CSV or NPY)")
parser.add_argument("--out", required=True, help="Output directory")
parser.add_argument("--labels", help="Label CSV path (optional)")
parser.add_argument("--topk", type=int, default=10, help="Ground truth")
args = parser.parse_args()
main(args.train, args.test, args.out, args.labels, args.topk)
Conversion Process Output:
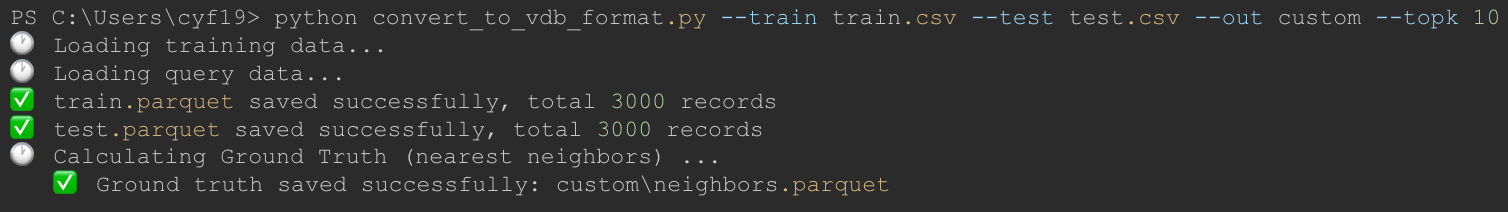
Generated Files Verification:

Custom Dataset Configuration
Navigate to the Custom Dataset configuration section in the web interface:
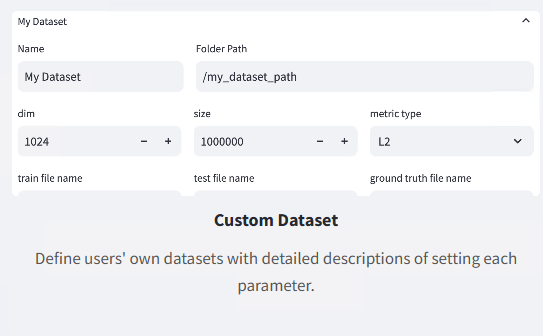
The configuration interface provides fields for dataset metadata and file path specification:
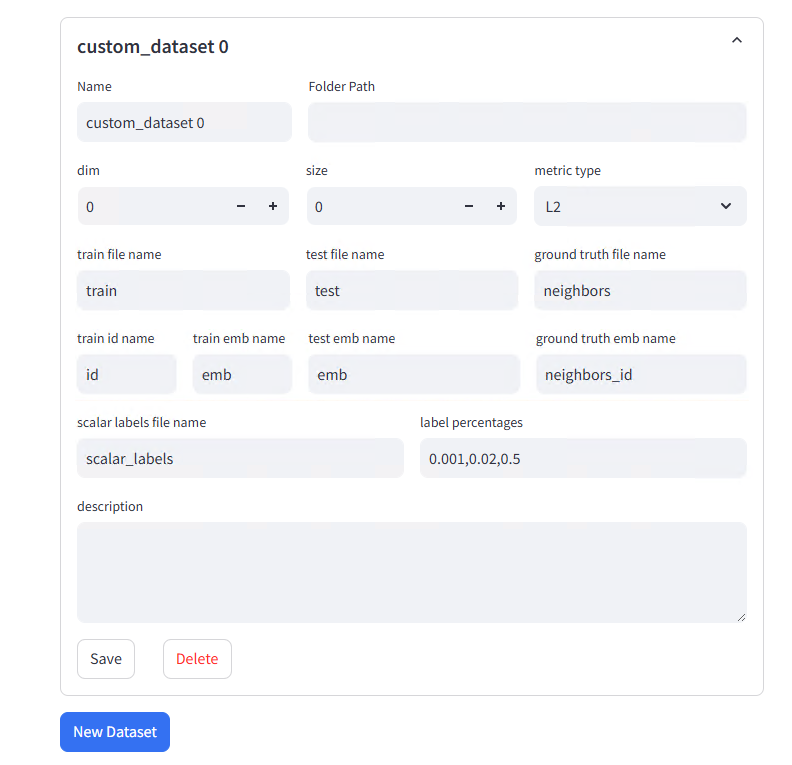
Configuration Parameters:
| Parameter Name | Meaning | Configuration Suggestions |
|---|---|---|
| Name | Dataset name (unique identifier) | Any name, e.g., my_custom_dataset |
| Folder Path | Dataset file directory path | e.g., /data/datasets/custom |
| dim | Vector dimensions | Must match data files, e.g., 768 |
| size | Vector count (optional) | Can be left empty, system will auto-detect |
| metric type | Similarity measurement method | Commonly use L2 (Euclidean distance) or IP (inner product) |
| train file name | Training set filename (without .parquet extension) | If train.parquet, fill train. Multiple files use comma separation, e.g., train1,train2 |
| test file name | Query set filename (without .parquet extension) | If test.parquet, fill test |
| ground truth file name | Ground Truth filename (without .parquet extension) | If neighbors.parquet, fill neighbors |
| train id name | Training data ID column name | Usually id |
| train emb name | Training data vector column name | If script-generated column name is emb, fill emb |
| test emb name | Test data vector column name | Usually same as train emb name, e.g., emb |
| ground truth emb name | Nearest neighbor column name in Ground Truth | If column name is neighbors_id, fill neighbors_id |
| scalar labels file name | (Optional) Label filename (without .parquet extension) | If scalar_labels.parquet was generated, fill scalar_labels, otherwise leave empty |
| label percentages | (Optional) Label filter ratio | e.g., 0.001,0.02,0.5, leave empty if no label filtering needed |
| description | Dataset description | Cannot note business context or generation method |
Save the configuration to proceed with the test setup.
Test Execution and Database Configuration
Access the test configuration interface:
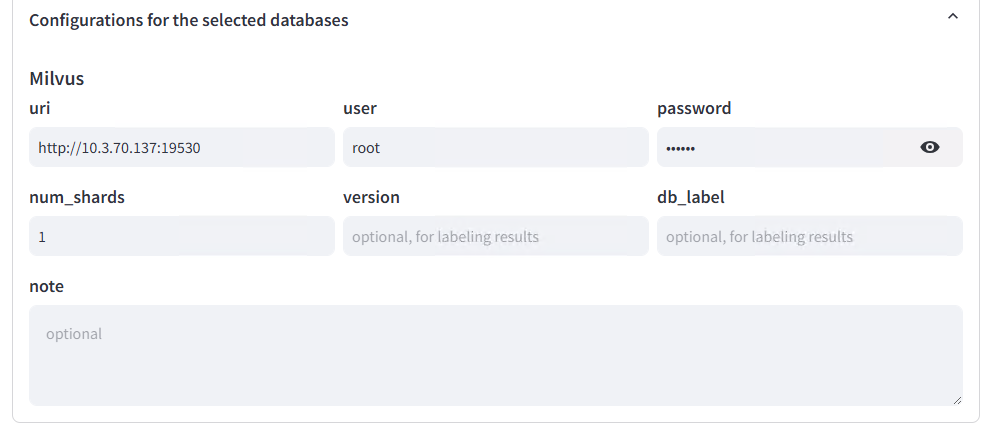
Database Selection and Configuration (Milvus as an Example):
Dataset Assignment:
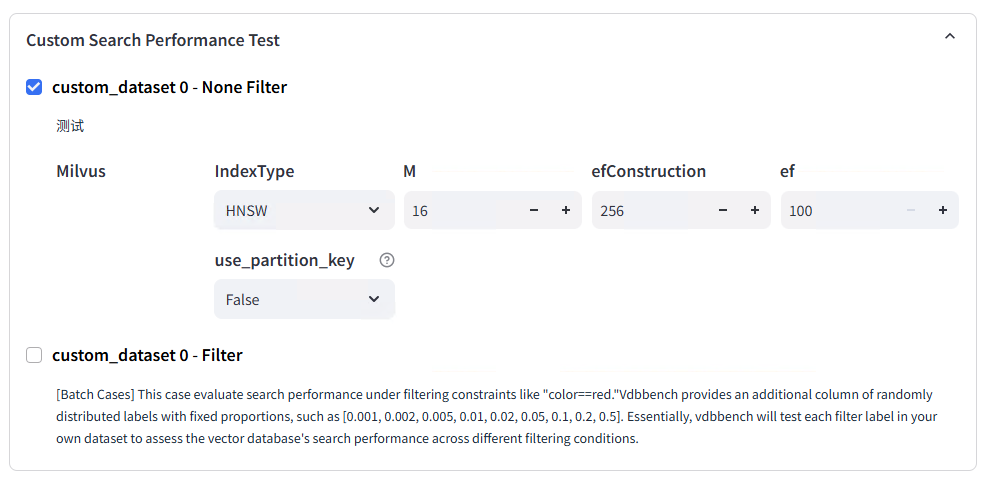
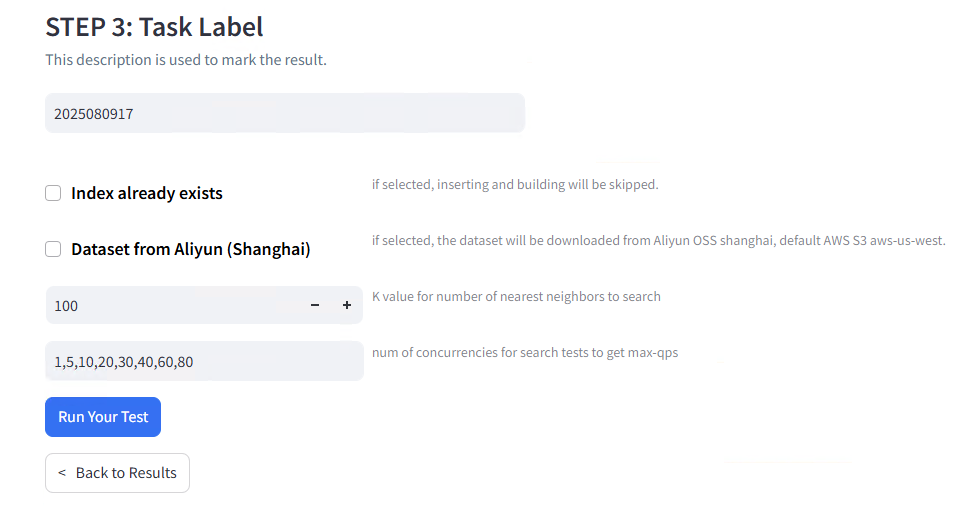
Test Metadata and Labeling:
Test Execution:
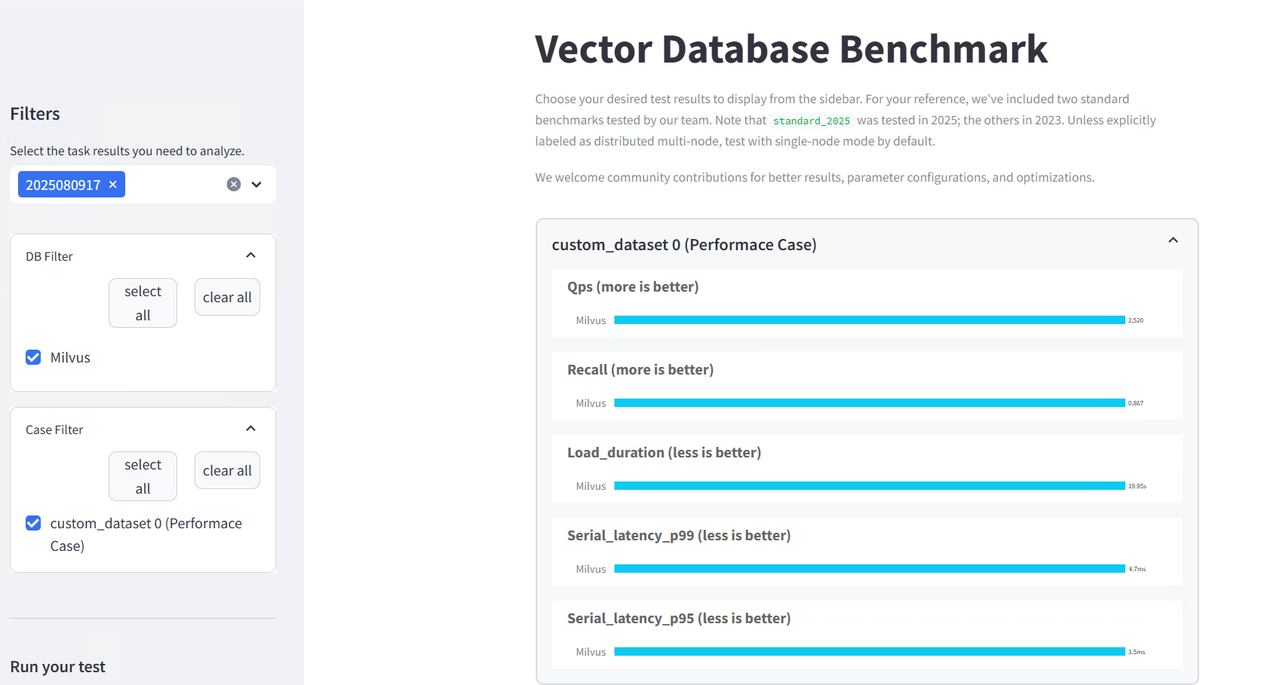
Results Analysis and Performance Evaluation
The results interface provides comprehensive performance analytics:
Test Configuration Summary
The evaluation tested concurrency levels of 1, 5, and 10 concurrent operations (constrained by available hardware resources), vector dimensions of 768, dataset size of 3,000 training vectors and 3,000 test queries, with scalar label filtering disabled for this test run.
Critical Implementation Considerations
Dimensional Consistency: Vector dimension mismatches between training and test datasets will cause immediate test failures. Verify dimensional alignment during data preparation to avoid runtime errors.
Ground Truth Accuracy: Incorrect ground truth calculations invalidate recall rate measurements. The provided conversion script uses FAISS with L2 distance for exact nearest neighbor computation, ensuring accurate reference results.
Dataset Scale Requirements: Small datasets (below 10,000 vectors) may produce inconsistent QPS measurements due to insufficient load generation. Consider scaling the dataset size for more reliable throughput testing.
Resource Allocation: Docker container memory and CPU constraints can artificially limit database performance during testing. Monitor resource utilization and adjust container limits as needed for accurate performance measurement.
Error Monitoring: VDBBench may log errors to console output that don’t appear in the web interface. Monitor terminal logs during test execution for complete diagnostic information.
Supplemental Tools: Test Data Generation
For development and standardized testing scenarios, you can generate synthetic datasets with controlled characteristics:
import pandas as pd
import numpy as np
def generate_csv(num_records: int, dim: int, filename: str):
ids = range(num_records)
vectors = np.random.rand(num_records, dim).round(6)
emb_str = [str(list(vec)) for vec in vectors]
df = pd.DataFrame({
'id': ids,
'emb': emb_str
})
df.to_csv(filename, index=False)
print(f"Generated file {filename}, {num_records} records total, vector dimension {dim}")
if
name
== "__main__":
num_records = 3000 # Number of records to generate
dim = 768 # Vector dimension
generate_csv(num_records, dim, "train.csv")
generate_csv(num_records, dim, "test.csv")
This utility generates datasets with specified dimensions and record counts for prototyping and baseline testing scenarios.
Conclusion
You’ve just learned how to break free from the “benchmark theater” that’s misled countless vector database decisions. With VDBBench and your own dataset, you can generate production-grade QPS, latency, and recall metrics—no more guesswork from decades-old academic data.
Stop relying on canned benchmarks that have nothing to do with your real workloads. In just hours—not weeks—you’ll see precisely how a database performs with your vectors, your queries, and your constraints. That means you can make the call with confidence, avoid painful rewrites later, and ship systems that actually work in production.
Try VDBBench with your workloads: https://github.com/zilliztech/VectorDBBench
View testing results of major vector databases: VDBBench Leaderboard
Have questions or want to share your results? Join the conversation on GitHub or connect with our community on Discord.
This is the first post in our VectorDB POC Guide series—hands-on, developer-tested methods for building AI infrastructure that performs under real-world pressure. Stay tuned for more!
- The Fix: Test with Your Data, Not Canned Benchmarks
- How to Evaluate VectorDBs with Your Custom Datasets with VDBBench
- Results Analysis and Performance Evaluation
- Supplemental Tools: Test Data Generation
- Conclusion
On This Page
Try Managed Milvus for Free
Zilliz Cloud is hassle-free, powered by Milvus and 10x faster.
Get StartedLike the article? Spread the word